Clonal evolution of leukemia is driven by different selection pressures operating on a heterogeneous population of cells. These pressures may include therapy-induced and immune-mediated effects which eradicate the majority of cells, but which simultaneously provide a selective advantage to a minor population of cells that is genetically or transcriptionally distinct and which grow out towards relapse.
We integrated single-cell RNA-seq, DNA-seq, ATAC-seq and long-read Oxford Nanopore sequencing of matched diagnosis and relapse samples of 20 pediatric AML patients with various genetic driver mutations representative of the mutational spectrum to map the genetic and transcriptional evolution of these cancers (Figure 1). We investigated whether and how differences in AML-initiating mutations impact on transcriptional programs in AML blasts and their surrounding microenvironmental cells and how the mutational and transcriptional features of these cells are impacted by cancer therapy. Specifically, we traced the progression of these traits from the point of diagnosis to the occurrence of relapse.
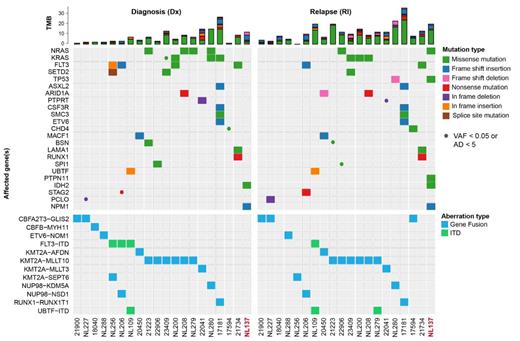
Our single cell RNA and ATAC data showed high patient-to-patient heterogeneity, with gene expression patterns that were largely conserved between diagnosis and relapse. Analyses of the differentially expressed genes and factorization into “metaprograms” revealed gene sets that were shared between relapse samples but could not be tied to interpretable common “biological signatures”. Exome-seq revealed highly significant changes in somatic variants, most notably in members of the RAS-signaling pathway (i.e., KRAS, NRAS, PTPN11, NF1 and CBL). Interestingly, pathogenic RAS mutations were often gained, but also lost, or switched between different RAS-pathway members, indicating that RAS-mutations are associated with extensive clonal evolution/rewiring. To unveil the effect of accumulation or loss of (sub)clones with RAS mutations on the transcriptional phenotype and epigenetic landscape of leukemic blasts, we performed targeted screening of somatic variants at single-cell resolution. To this aim, we combined long-read nanopore sequencing with 10x Genomics single-cell technologies, enabling us to directly link genetic, transcriptomics and epigenetic changes in distinct (sub)clones.
Taken together, we successfully reconstructed patient-specific genetic and transcriptional evolutionary trajectories in pediatric AML. This knowledge will be essential for our understanding of AML relapse and therapy resistance.
Disclosures
Heidenreich:Roche: Research Funding; Syndax: Research Funding. Zwaan:Pfizer: Other: Institutional support for clinical trials; Abbvie: Other: Institutional support for clinical trials; Takeda: Other: Institutional support for clinical trials; Jazz: Other: Institutional support for clinical trials; Kura: Other: Institutional support for clinical trials; Gilead: Other: Institutional support for clinical trials; Daiichi Sankyo: Other: Institutional support for clinical trials; Kura Oncology: Consultancy; BMS: Consultancy; Novartis: Consultancy; Gilead: Consultancy; Incyte: Consultancy; Syndax: Consultancy; Novartis: Membership on an entity's Board of Directors or advisory committees; Sanofi: Membership on an entity's Board of Directors or advisory committees; Incyte: Membership on an entity's Board of Directors or advisory committees; ITCC Hem Malignancies Committee: Other: Leadership or fiduciary role in other board, society, committee, or advocacy group, paid or unpaid; Chair MREC Utrecht: Other: Leadership or fiduciary role in other board, society, committee, or advocacy group, paid or unpaid; Chair Dutch MREC Society: Other: Leadership or fiduciary role in other board, society, committee, or advocacy group, paid or unpaid.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal